Single Cell Transcriptomics
Kerry Cobb
Objectives & Learning Goals for Workshop
- HPC Usage
- Singe Cell Transcriptome Concepts
- Technical Details
Objectives & Learning Goals for Workshop
- HPC Usage
- Accessing remote HPC
- Submitting jobs
- Basic linux usage
Objectives & Learning Goals for Workshop
- Singe Cell Transcriptome Concepts
- Sample preparation
- Read mapping
- Quality control
- Analysis
Objectives & Learning Goals for Workshop
- Technical Details
- Available software
- Scripts to run analyses
Objectives & Learning Goals for today
- Introduction to single cell transcriptomics
- Overview of sample preparation, library construction, and sequencing
- Initial data processing
Single Cell Transcriptomics
- Why do single cell transcriptomics?
- Characterize heterogeneity among cells within cell population
- Identify rare cell types
- Explore interaction and communication among cells
- Trace cell lineages during development
Bulk vs Single Cell
- Bulk transcriptomics
- Measures average expression across all cells in a sample
- Cannot detect heterogeneity among cells
- Higher gene coverage
- Single cell transcriptomics
- Measures expression in individual cells
- Can detect heterogeneity among cells
- Lower gene coverage
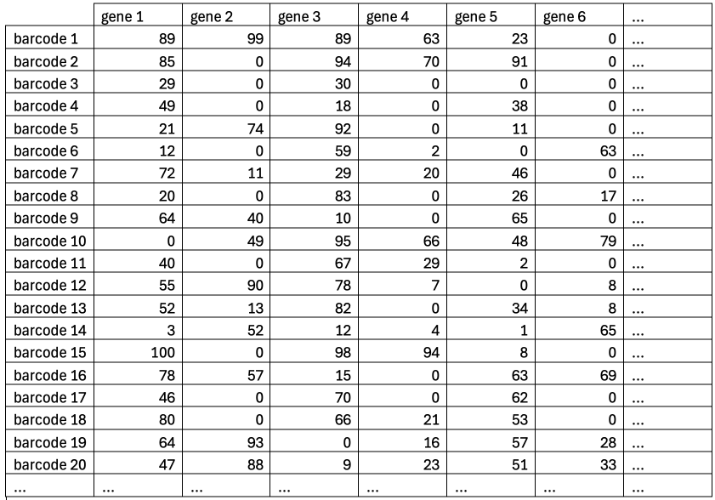
Single Cell Transcriptomics Workflow
- Tissue preparation
- Single cell isolation

- Library preparation

- Sequencing

- Data processing & analysis
1. Tissue Preparation
- Critical Step (Garbage in, garbage out)!
- Many different methods
- Details beyond the scope of this workshop
- Cell selction / enrichment
- Sample clean-up
- Considerations for each method w/ regards to analysis
- Expected cells
- Quality of cells
- Batch effects
1. Tissue Preparation
- Take steps to remove batch effects & confounding variables
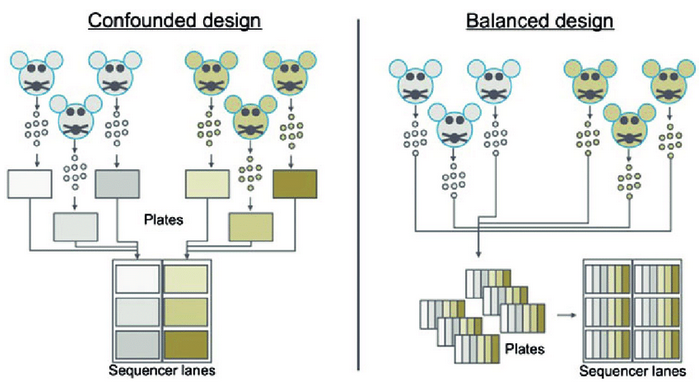
- Consider replication if feasible
2. Single Cell Isolation
Svensson et al. 2017
2. Single Cell Isolation
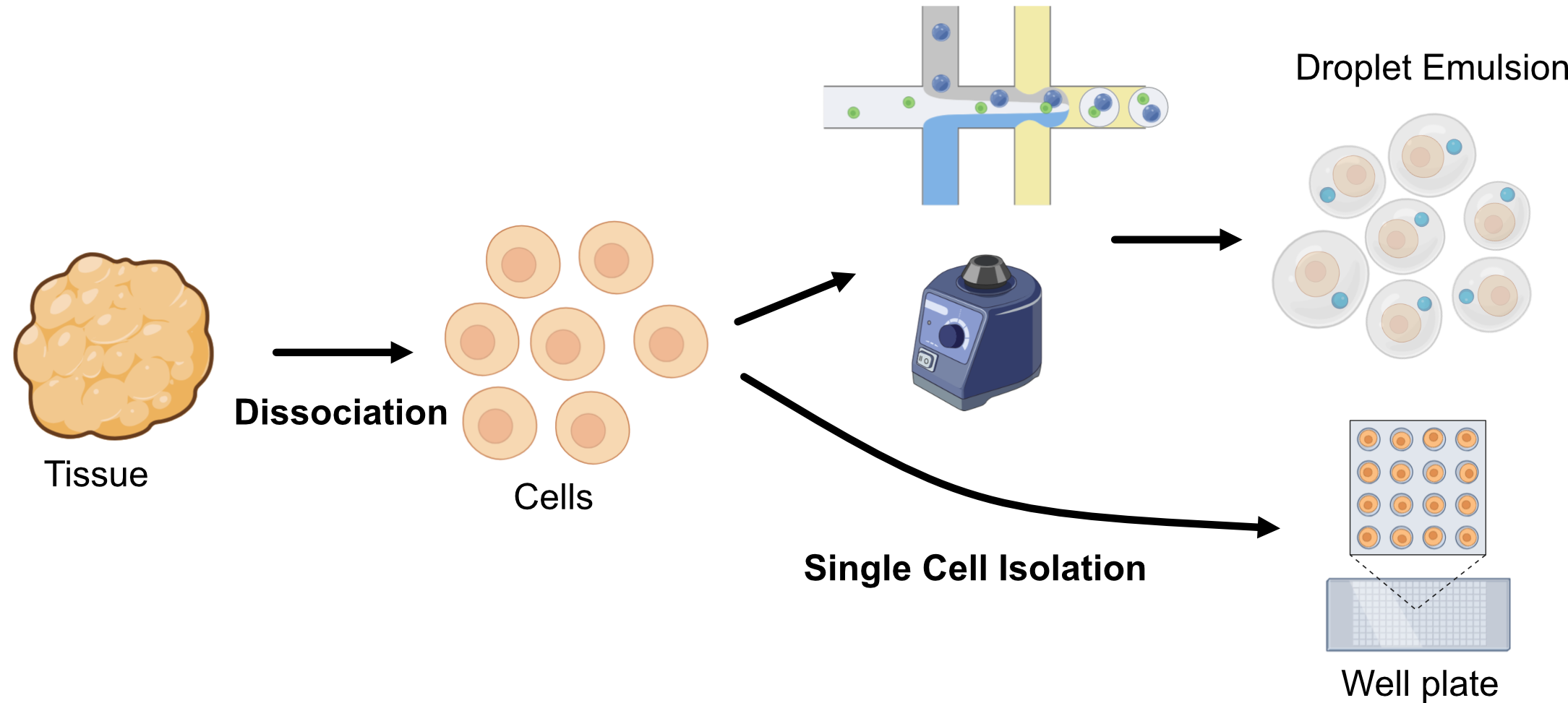
Multiple methods broadly categorized:
- Emulsion based
- Plate based
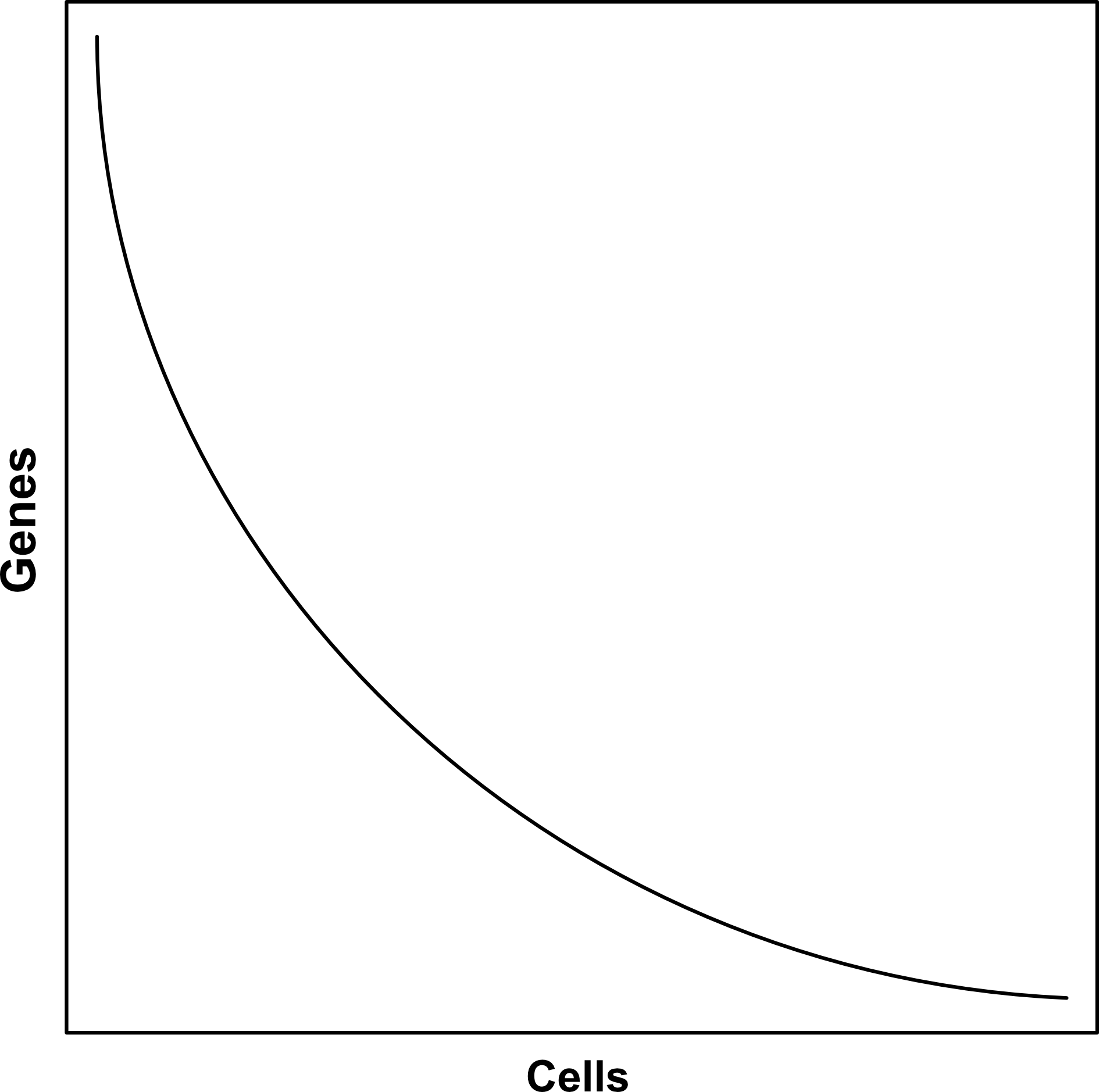
Single Cell Isolation - Trade off between cells and genes
- Depends on:
- Method used
- Choices made in implementation of method
- Required number of cells increases with complexity of sample
- Sample can often be resequenced
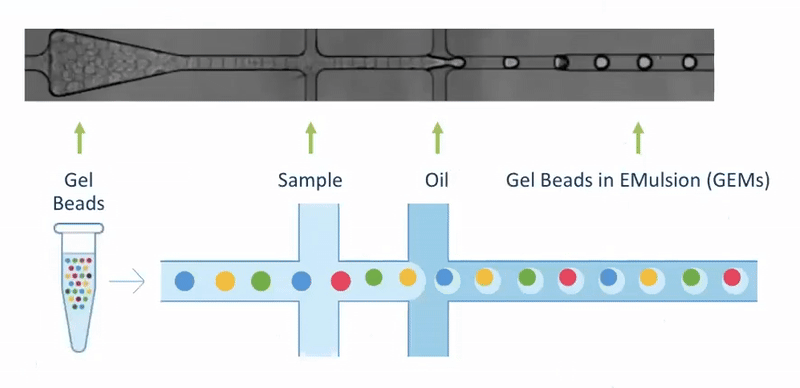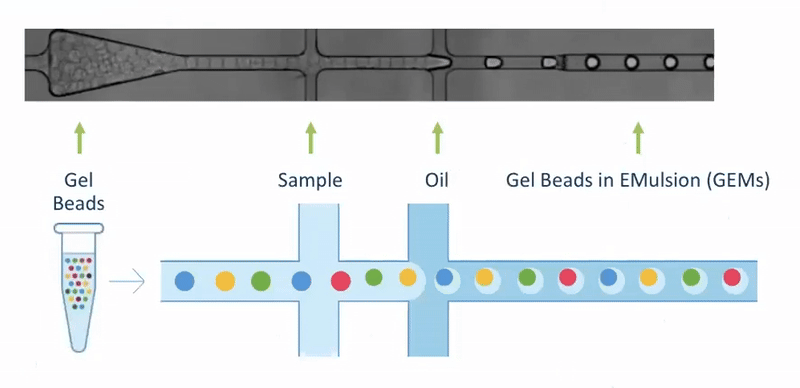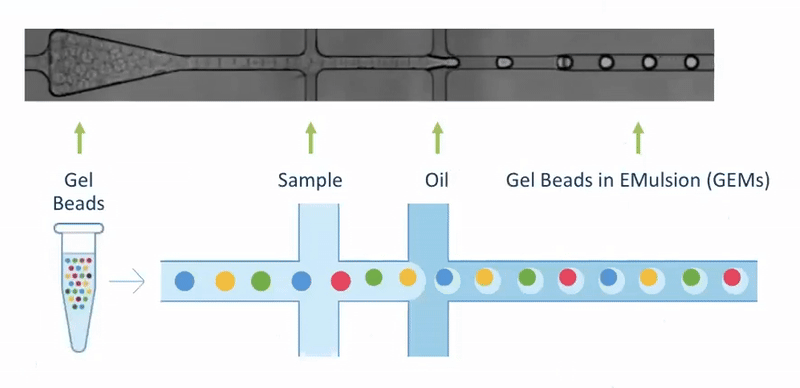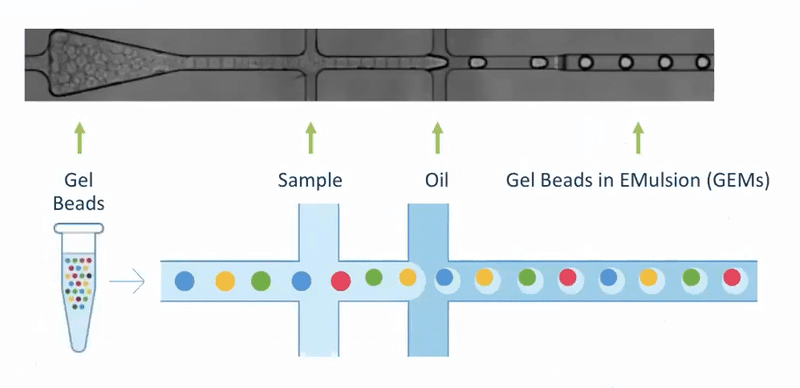
Single Cell Isolation - 10X Genomics
- By far the most popular approach
- Source of all workshop data

Single Cell Isolation - 10X Genomics
Single Cell Isolation - Illumina single cell 3’ RNA
- Very recent offering
- Formerly known as PIPSeq
- Looks very promising, several UConn & UCHC labs planning to adopt
Clark et al. 2023
Single Cell Isolation - Combinatorial Barcoding
Parse Biosciences Evercode
- Cells serve as reacion vessel
- Serial splitting, barcoding, and pooling results in each cell having a unique combination of barcodes
- C(96,4) = 3,321,960
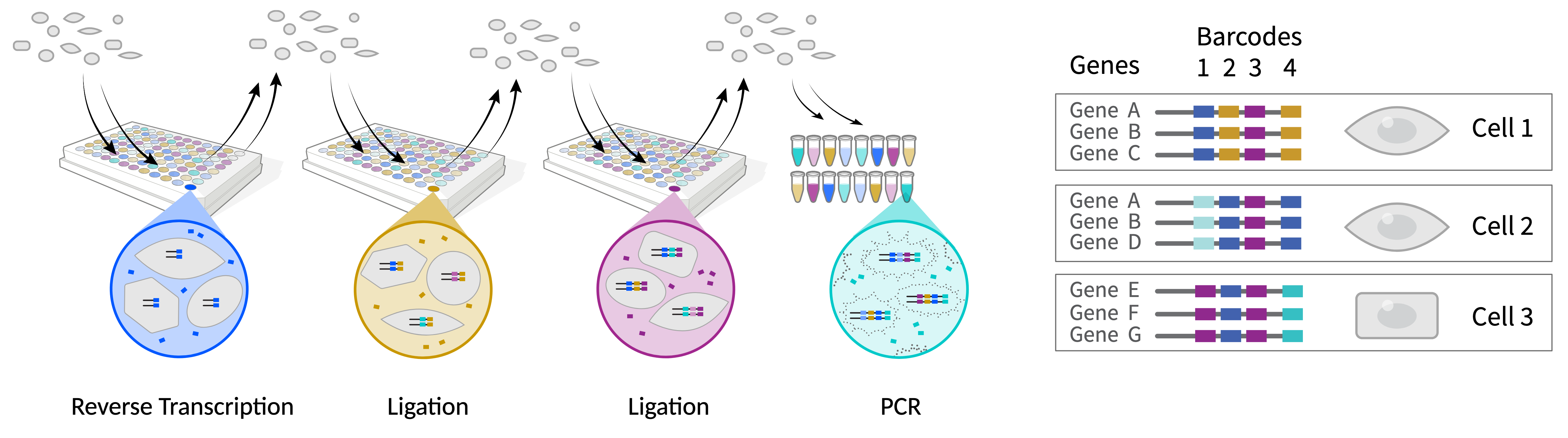
Tran et al. 2022
Single Cell Isolation - Combinatorial Barcoding
Scale Biosciences Quantum Barcoding
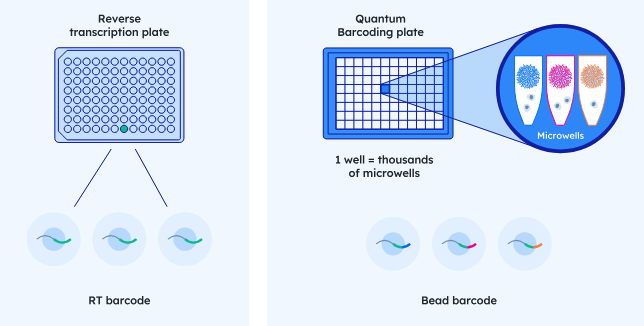
Single Cell Isolation - SMART-SEQ
- Plate-based
- Much greater cost per cell
- Can assay full transcripts
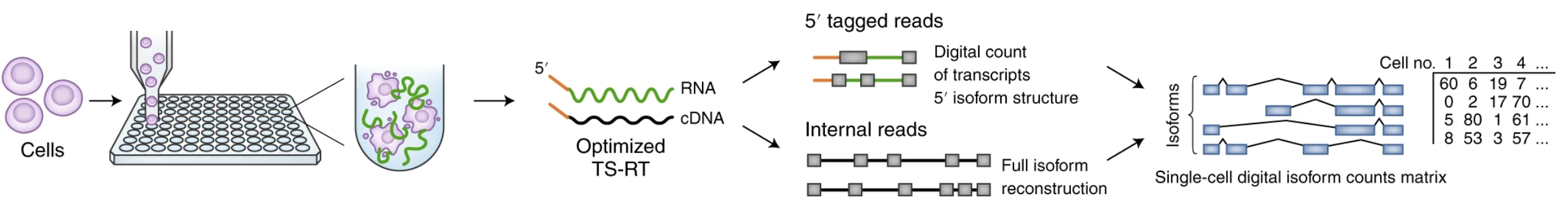
Macosko 2020
Single Cell Isolation - Others
- Honeycomb HIVE
- Singleron
- Asteria
- Fluent
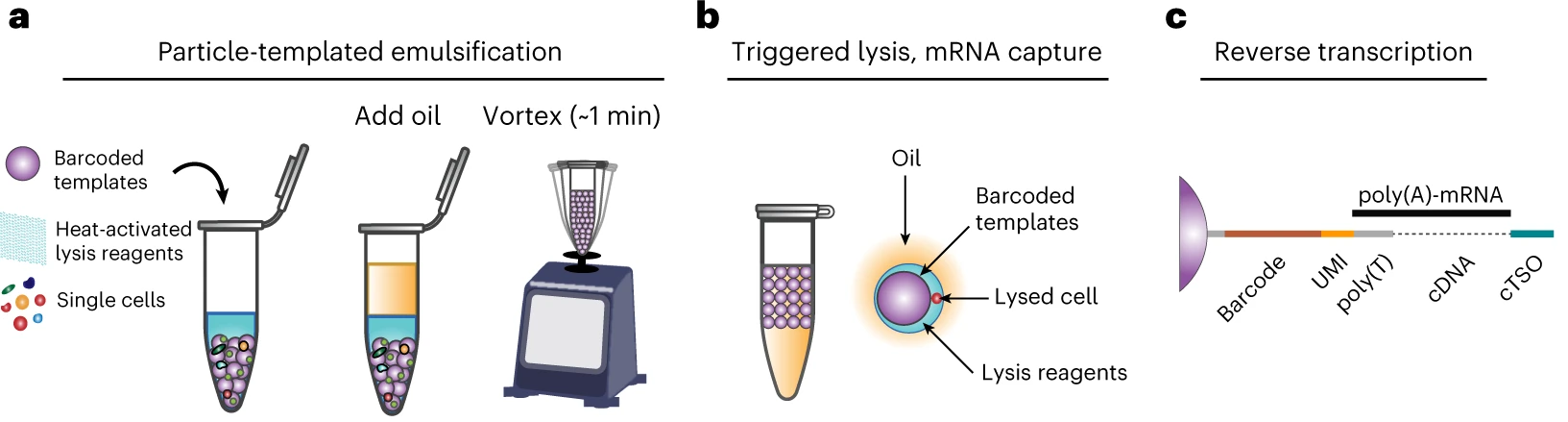
3. Library Preparation
- Convert RNA to cDNA
- Reverse transcription
- Fragmentation
- Barcode ligation
- Unique molecular identifiers (UMIs)
- Cell barcodes
- Sequencing adapter ligation
- Amplification
- Remove contimating RNA and cell debris
10X Library Preparation


10X RNA Capture
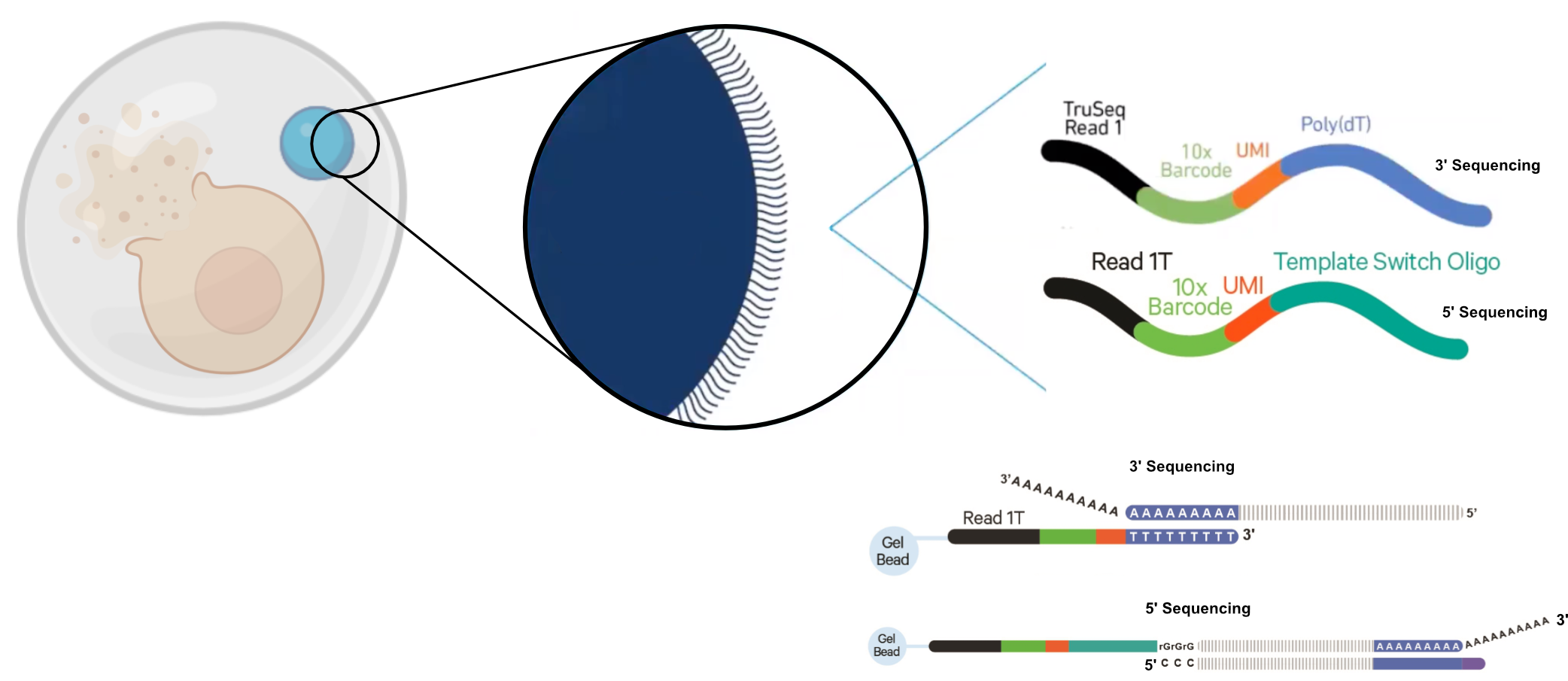
Adapted from 10X Genomics
10X 3` vs 5` libraries


- Refers to the end of an RNA molecule that is sequenced
- 3` came first
- 5` permits identification of T-cell and B-cell receptor sequence
- Sensitivity is comparable according to 10X
10X 3` Capture

10X Genomics
10X 5` Capture
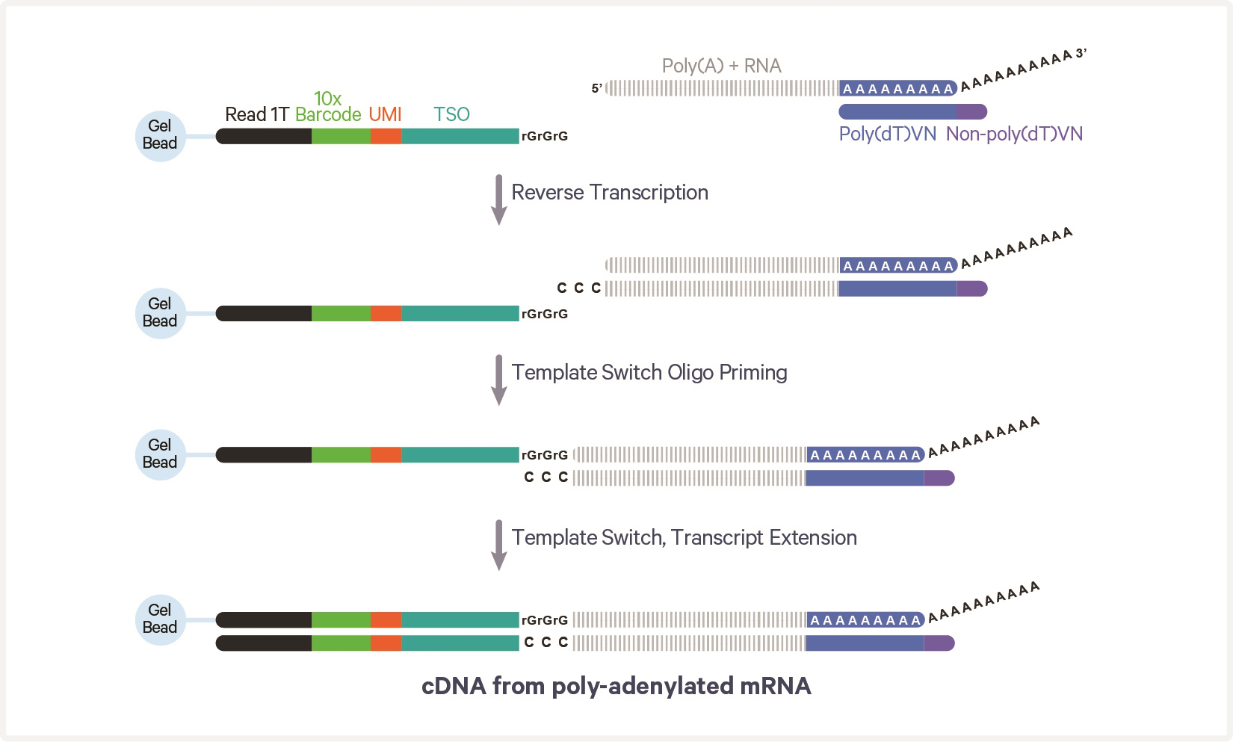
10X Genomics
10X 3’ Library construction
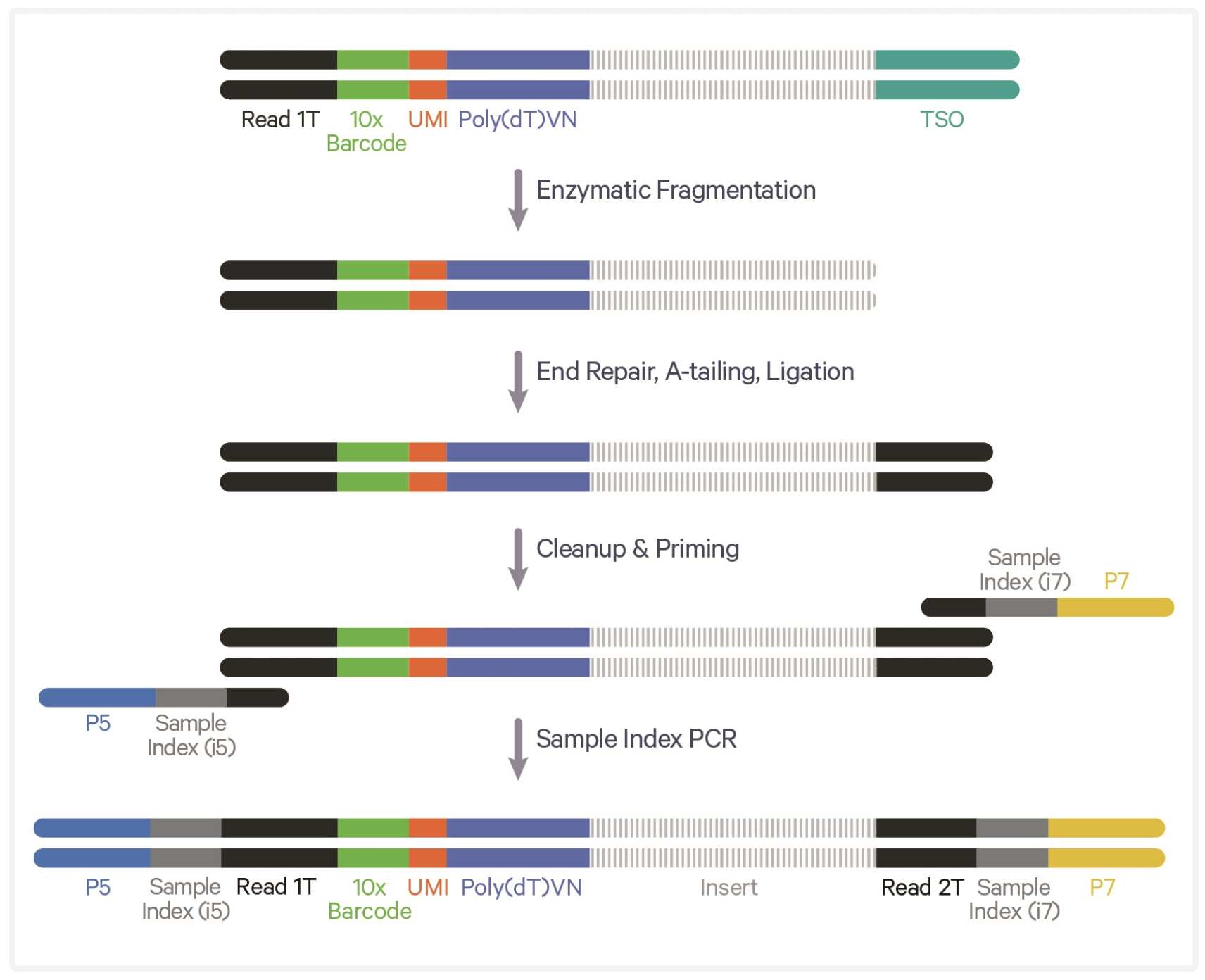
10X Genomics
10X 5’ Library construction
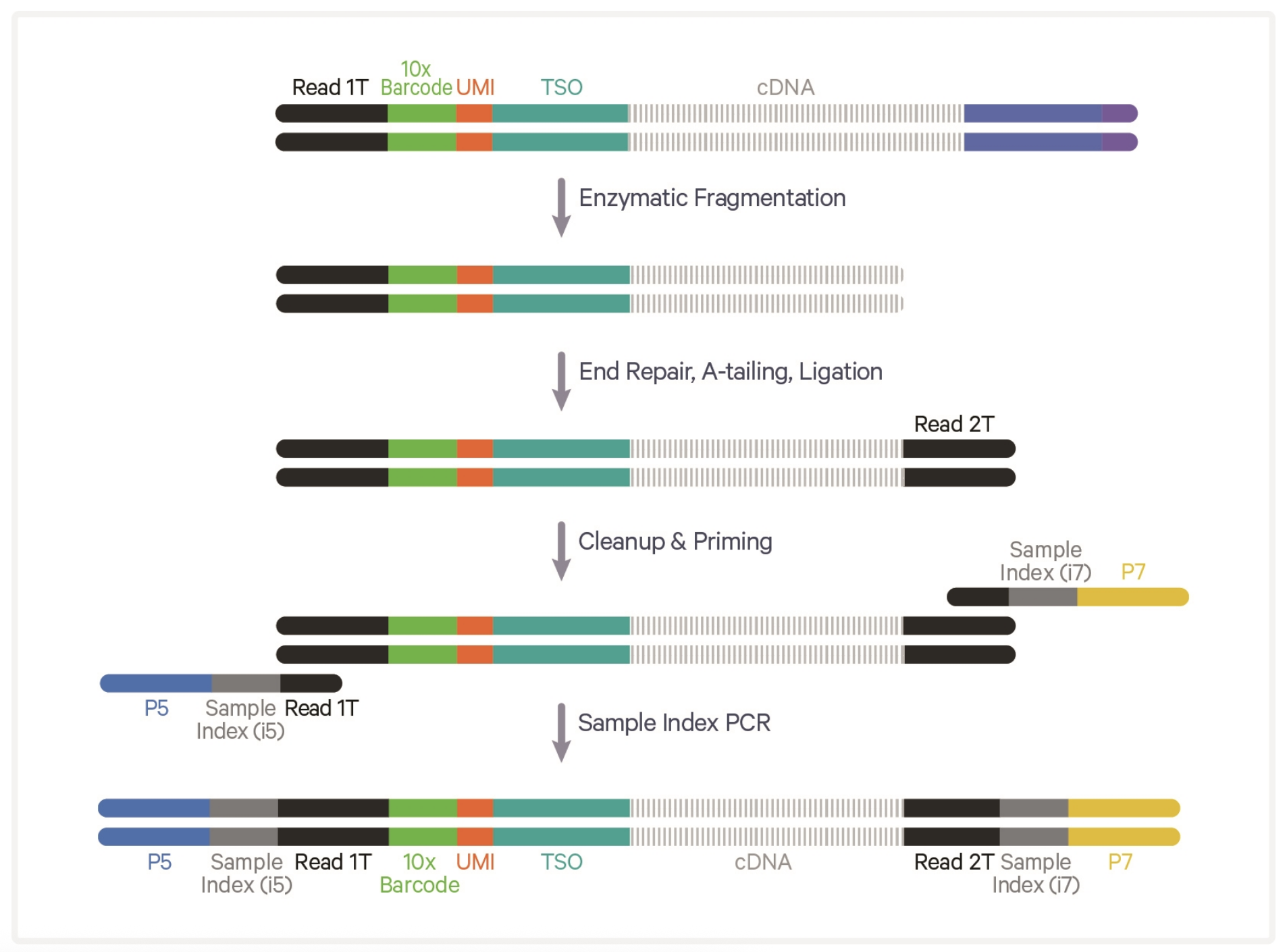
10X Genomics
Constructing Libraries from mRNA
- 10X is most common
- Multiple versions
- Other methods exist
- Method used can have important implications for analysis
- Index hopping
- PCR duplicates
- Biased amplification
- Biased capture?
Unique Molecular Identifiers (UMIs)

- Permit identification of PCR duplicates
- Results in more accurate estimate of expression
- 10X Genomics uses a random 12 nt sequence on each oligo
4. Sequencing
- Typically done with Illumina short reads
- Focus of workshop
- Could be done with any sequencing platform
- Each may require special considerations
- Read length
- Error rate
- Technical artifacts/biases
- Each may require special considerations

Multiplexing samples
- Samples can be multiplexed
- Two approaches
- Cell labelling
- SNP variation
- Can reduce impact of batch effects
- Cell labelling permits overloading
- Reduces empty droplets, doublets easy to identify
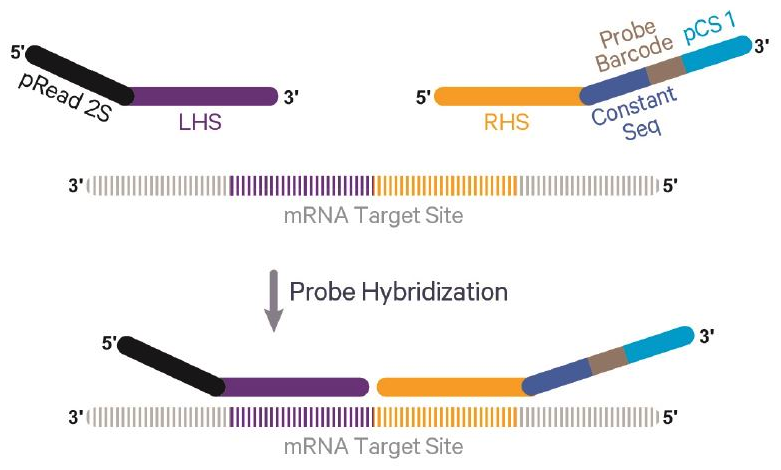
Probe capture
- Generally
- RNA captured by hybridization with probes rather than poly A tail
- Permits sequencing of formalin fixed tissue and low quality tissues
- Adds additional cost
- Limited to humans and mice
10X Genomics
Single-cell vs Single-nuclei
Single Cell
- Can detect transcripts in the cytoplasm as well as the nucleus
- Typically want to use fresh cells
- Signal more prone to perturbation caused by tissue processing
Single Nuclei
- Can detect more unprocessed mRNA containing introns
- Cannot detect transcripts in the cytoplasm making it unsuitable for some investigations
- Can be use with preserved cells and difficult to dissociate cells
5. Data Processing
- Up next!